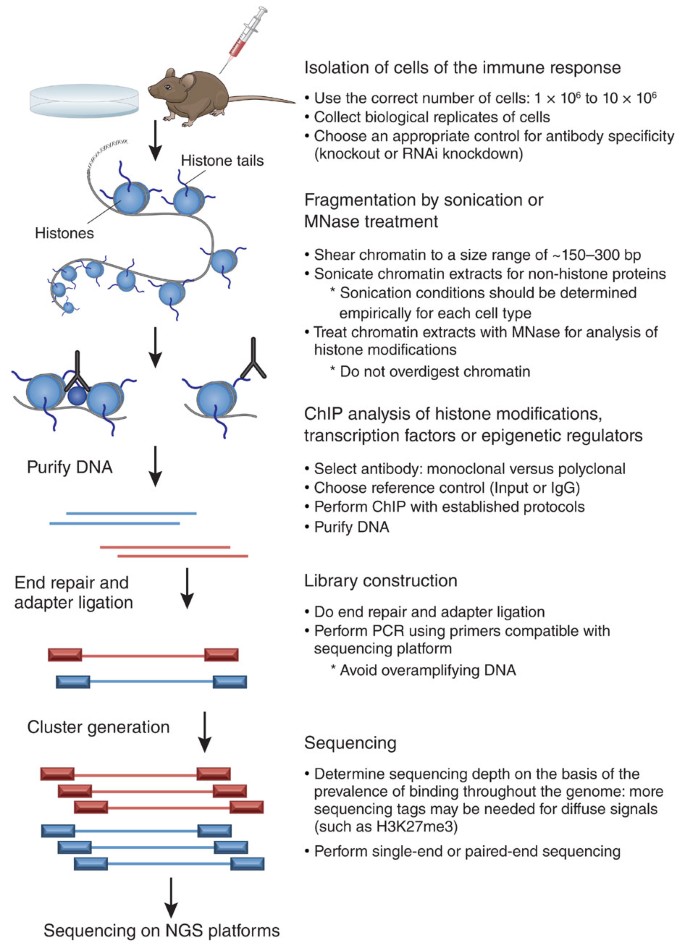
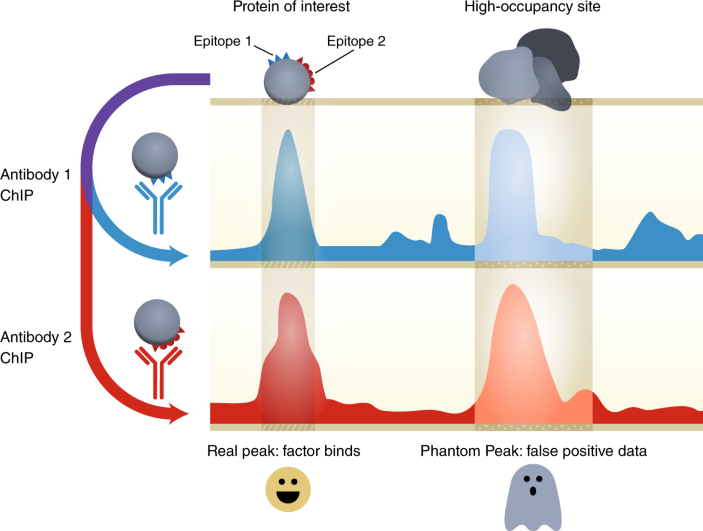
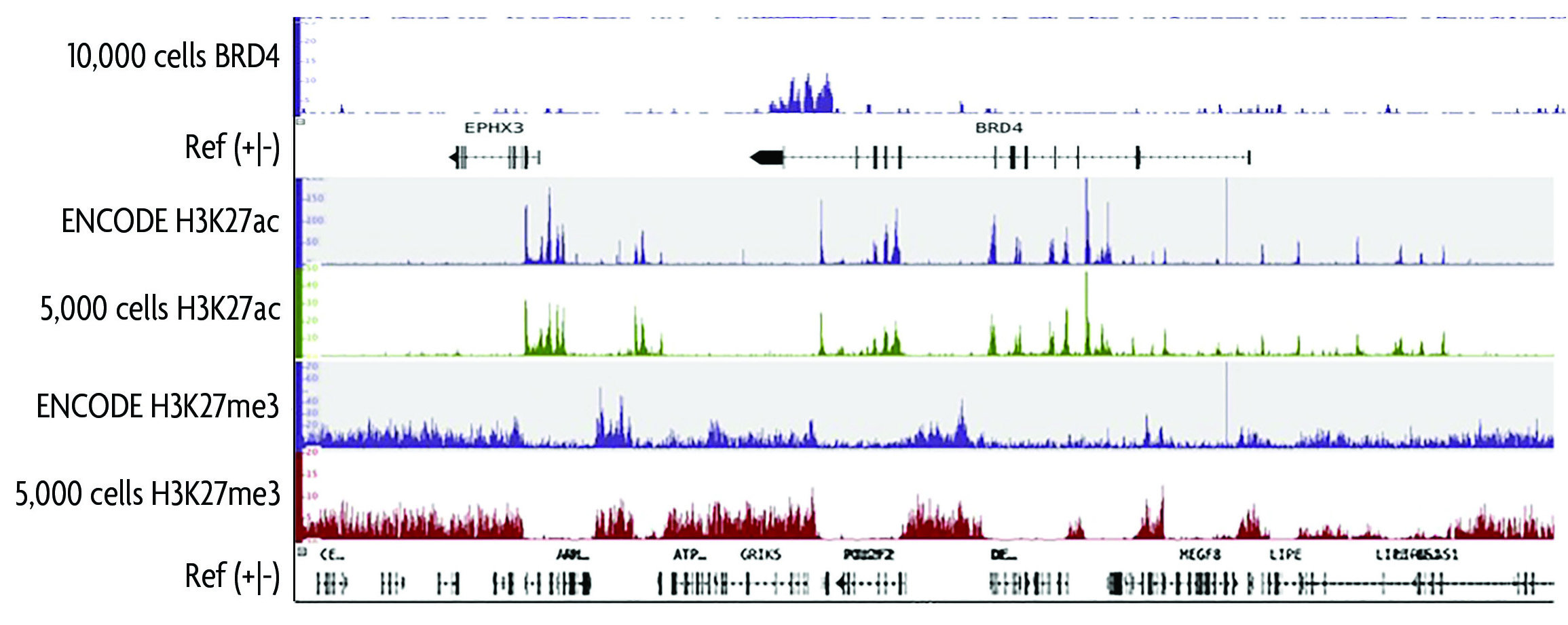
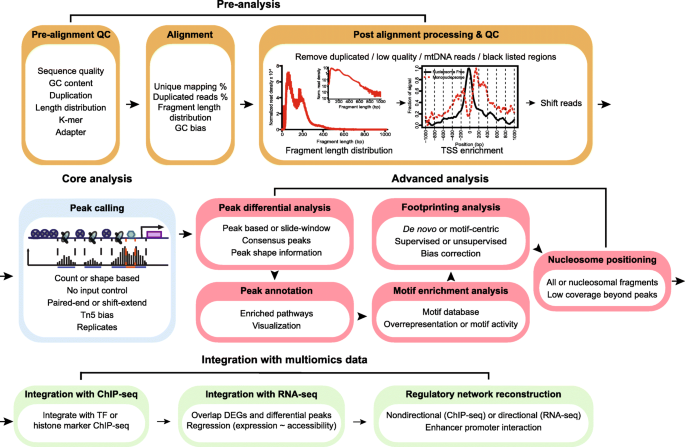
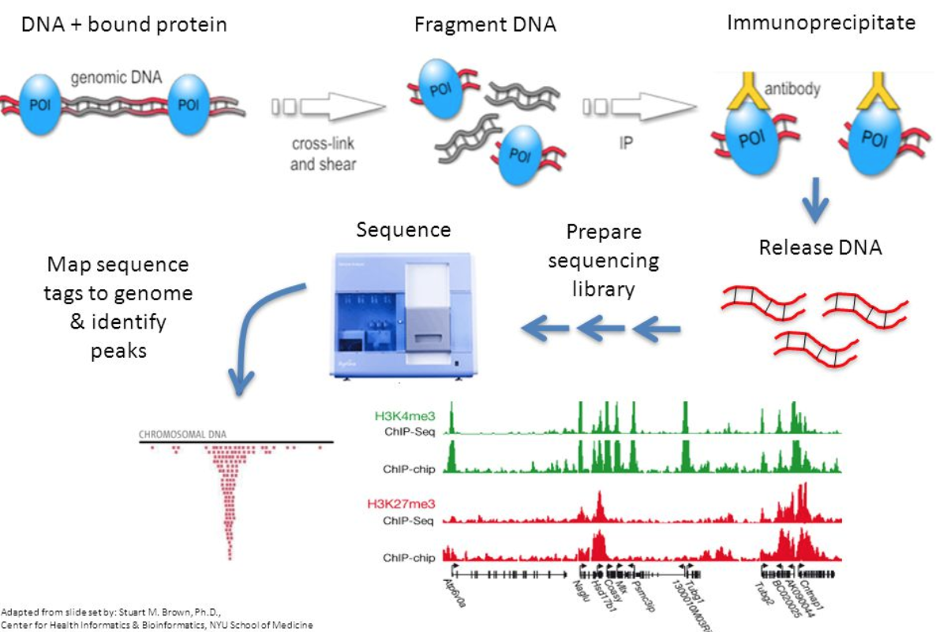
Protocol to apply spike-in ChIP-seq to capture massive histone acetylation in human cells - ScienceDirect

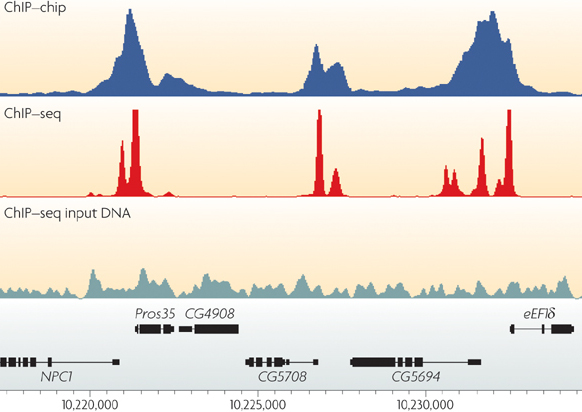
Chromatin immunoprecipitation and multiplex sequencing (ChIP-Seq) to identify global transcription factor binding sites in the nematode Caenorhabditis elegans. | Semantic Scholar

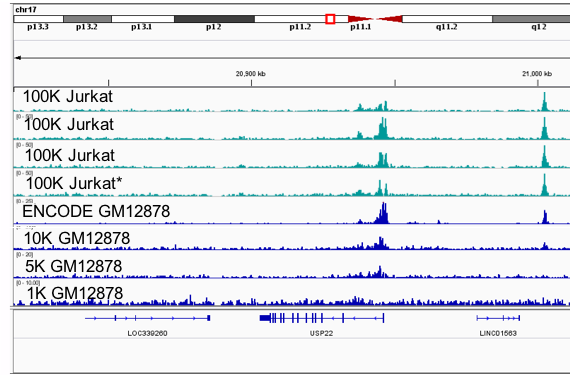
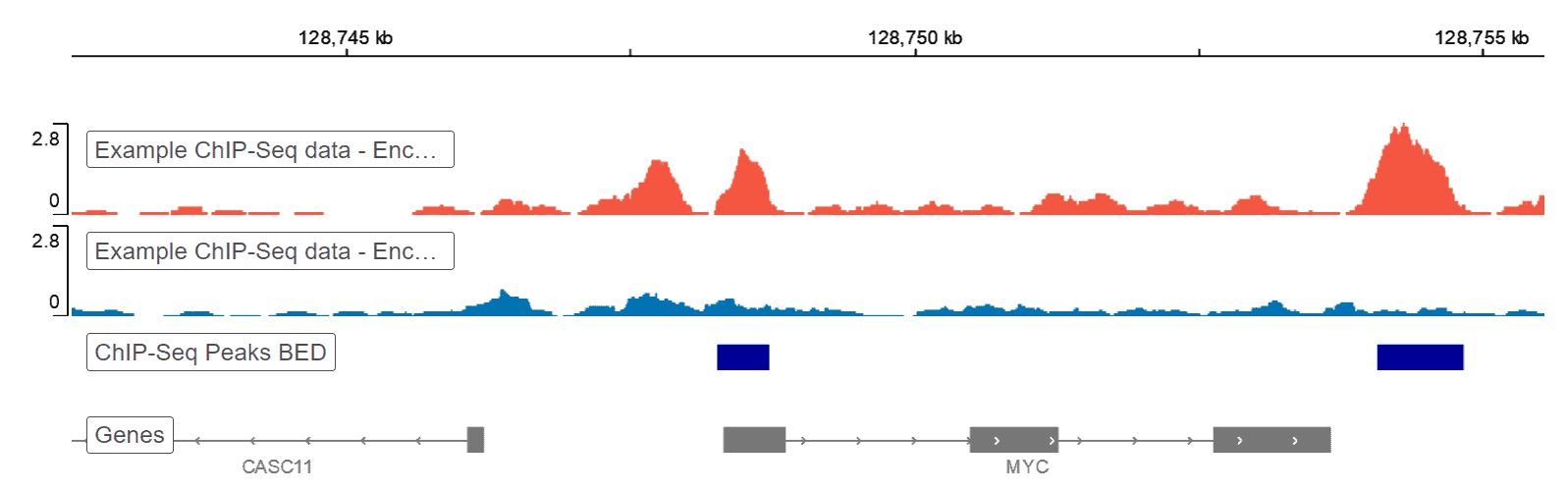
Quality control and data metrics of ChIP-seq data. (A) Example genomic... | Download Scientific Diagram

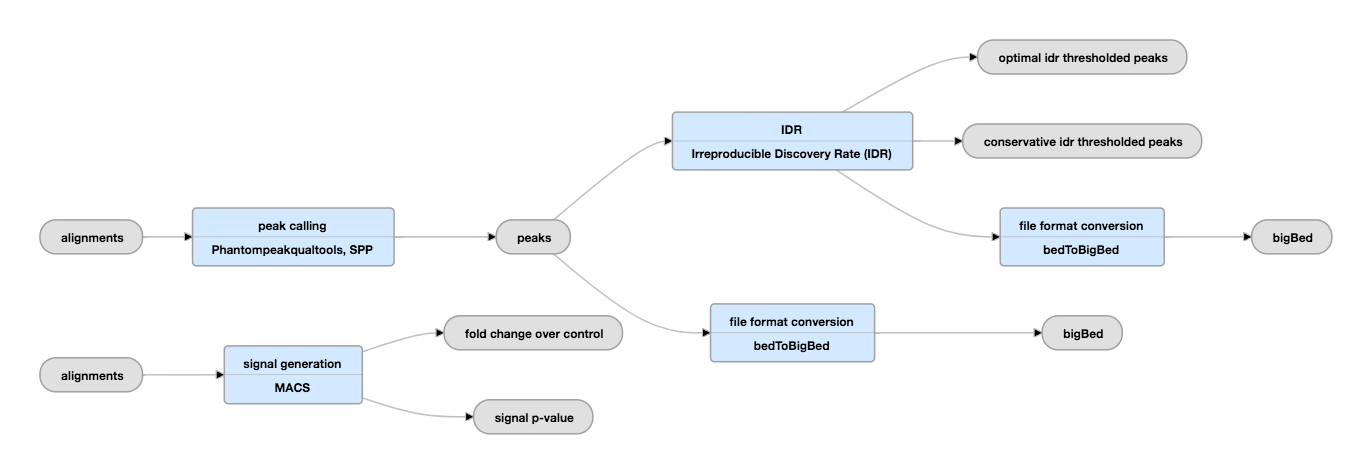
Troubleshooting the ChIP-seq workflow: From sequence reads to peak calls | Introduction to ChIP-Seq using high-performance computing